Overview
The problem of fMRI pre-processing is profound and urgently needs rigorous attention to ensure the continuing high quality of basic and clinical neuroimaging research in the future, particularly given the propensity for ever-more complex derivation of functional-connectivity based biomarkers in resting-state fMRI. Extracting BOLD signal from fMRI data is challenging due to many reasons including but not limited to: high scanner and thermal noise, irregular sampling in interleaved slice acquisition, physiological noise, motion and artifactual related contamination, and inter-subject variability. Even more challenging is the interaction between these phenomena that somehow makes their optimal correction almost impossible. For instance, the interaction between slice timing and motion prevented the field to be able to consent on the order of which their corrections should be applied. Here in QNL we aim at using signal and image processing techniques to optimally extract the BOLD signal from the fMRI data. We have multiple projects tackling different pre-processing pipeline.
Optimal Recovery of BOLD signal from Interleaved fMRI Data
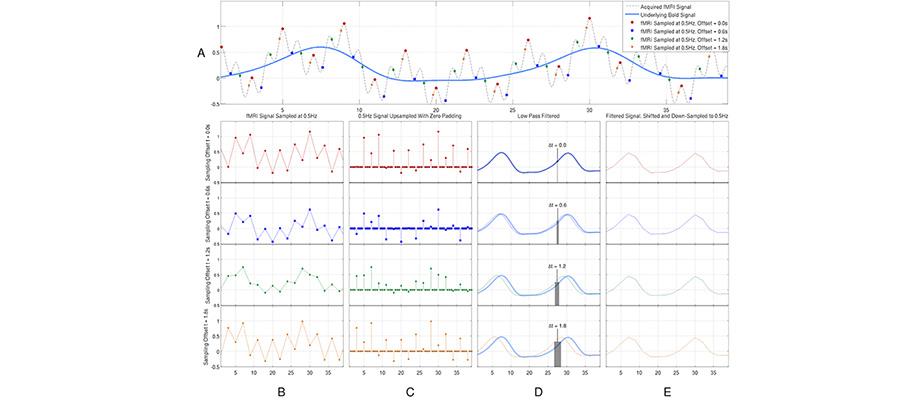
We have developed an optimal technique to extract the BOLD signal from interleaved fMRI data. In his project we use a simple signal reconstruction method that has been used in the field of signal processing for many years to correct of irregular sampling problem in interleaved slice acquisition. We have shown the superiority of this optimal method in compare to the existing methods and it’s interactions with other fMRI artifacts and processing steps. The software is implemented in C++ and is available for sharing online through github software development and sharing database.
Region-based Spatial Normalization
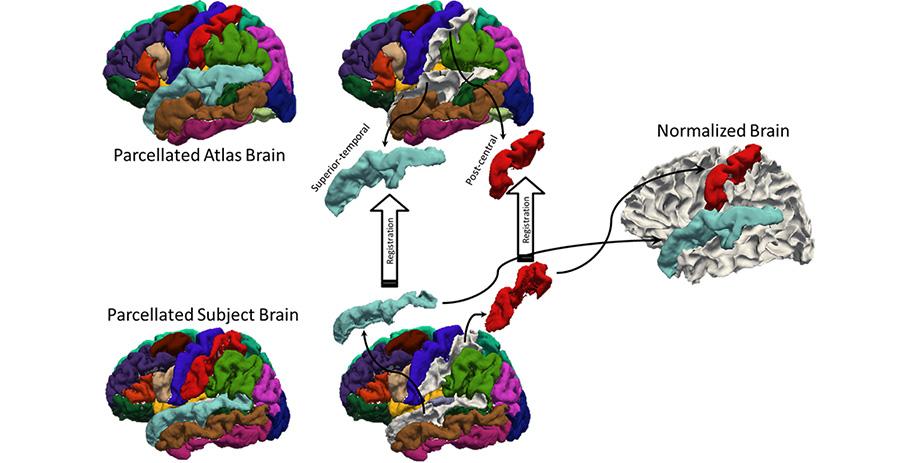
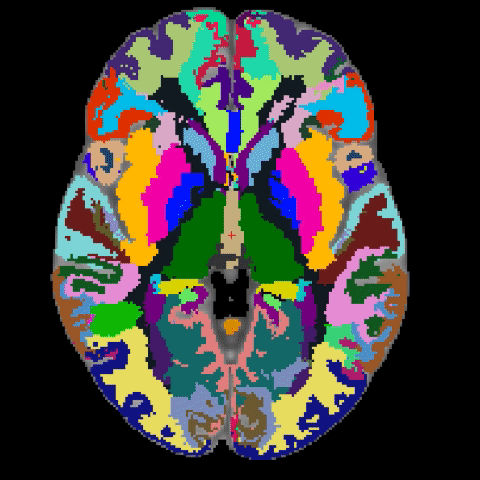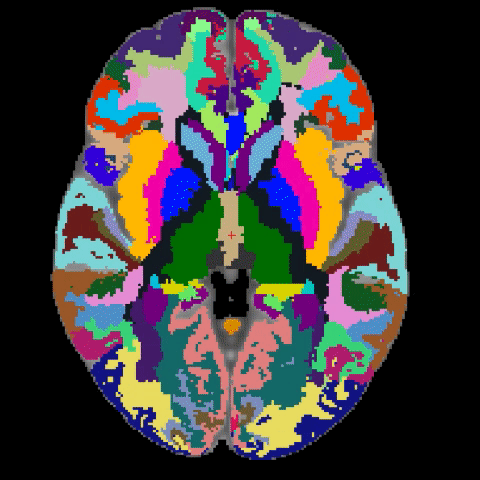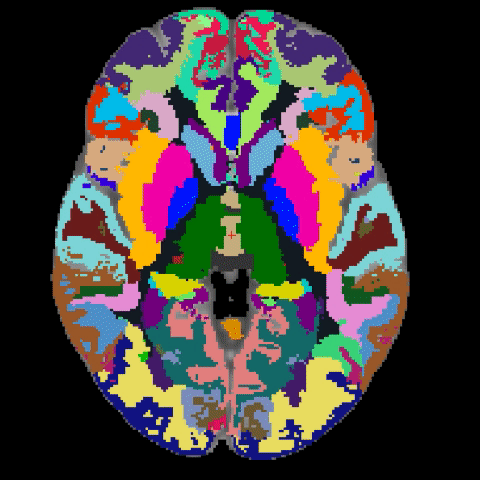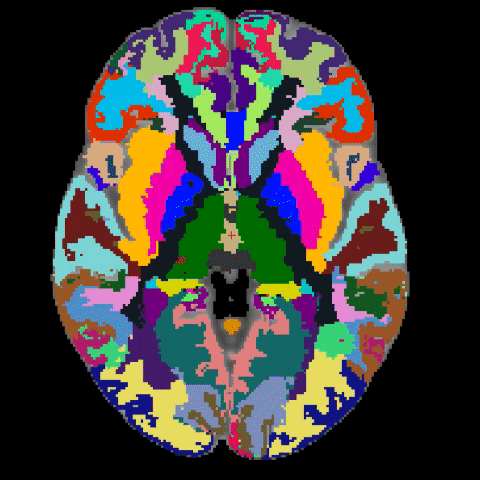
A longstanding problem in functional neuroimaging studies of cognitive aging is that age-related changes in brain morphology make it difficult to co-register brains, a key step for studies comparing task-related activation in young and old groups. To demonstrate the severity of the problem, the below video shows 29 participants’ brains after spatial normalization. The corresponding regions are shown with the same color. The video clearly shows the magnitude of the variability for each voxel between subjects. To address this issue, we are developing a region-based spatial normalization (RBSN) technique that will increase the accuracy of the fMRI data localization. The prevailing spatial normalization method tries to align all regions of the brain at once. RBSN, on the other hand, aligns each neuroanatomical region of the human brain independently. RBSN will thus provide more accurate localization of activation and ensure that group analyses test the same brain area in each study participant. Better between-participant registration also provides additional statistical power to detect activation in regions that may not have reached the significance level using prevailing methods (i.e.it will reduce type II error). It may also rule out previously noted areas of activation that were detected for artifactual reasons (i.e., it will reduce type I error).
Related Publication
D.B. Parker, X. Liu, Q. R. Razlighi, “Optimal slice timing correction and its interaction with fMRI parameters and artifacts,”Medical Image Analysis, In Press, Aug. 2016.
D.B. Parker, R.T. Gerraty, Q.R. Razlighi, “Optimal signal recovery from interleaved FMRI data,” IEEE international Symposium on Biomedical Imaging, Brooklyn, New York, 2015.
Software Released
Version 1.3 is the most updated and robust package which can be downloaded from github using the link below. The binaries for CentOs 6.5 is also available. Running the code with no input will print out the help which should be self sufficient for its execution. Please report the bugs to Dr. Razlighi for investigation.

